In Vitro Modeling of Gut Microbiomes for ExploringHost-Microbe Interactions
Overview
This research focuses on developing and applying in vitro gut microbiome models as a platform to study host-microbe interactions, particularly those linked to inflammation. These controlled model systems provide a powerful tool to dissect how microbial communities respond to environmental factors, such as nutrient availability, and how these interactions shape their composition, metabolic activity, and functional capacity. By enabling a mechanistic investigation of microbial dynamics, this approach would help reveal critical insights into their contribution to inflammatory pathways and related human diseases. Besides, we aim to leverage these in vitro microbial models to identify functional microbial traits that can serve as targets for precision microbiome editing, ultimately advancing therapeutic strategies. This work looks to uncover useful insights that pave the way for personalized approaches to microbiome modulation. To achieve this, we employ an integrative approach that combines in vitro anaerobic cultivation, genome-resolved metagenomics, bioinformatics, cell-based assays, molecular biology techniques, and metabolomics. This multi-faceted methodology allows us to recreate, characterize, and analyze microbial ecosystems, their metabolic outputs, and their interactions with host systems.

Research Areas and Approaches
From Gut to Lungs: The Infant Gut Microbiome Role in Inflammation and Asthma Development
Within this broader framework, one current research project investigates the infant gut microbiome and its role in inflammation and asthma development. Early-life gut microbiomes are crucial for immune system development, and their composition and functionality are shaped by various factors such as diet, environment, and microbial interactions. Disruptions in these early microbial ecosystems have been linked to an increased risk of asthma and allergic diseases.
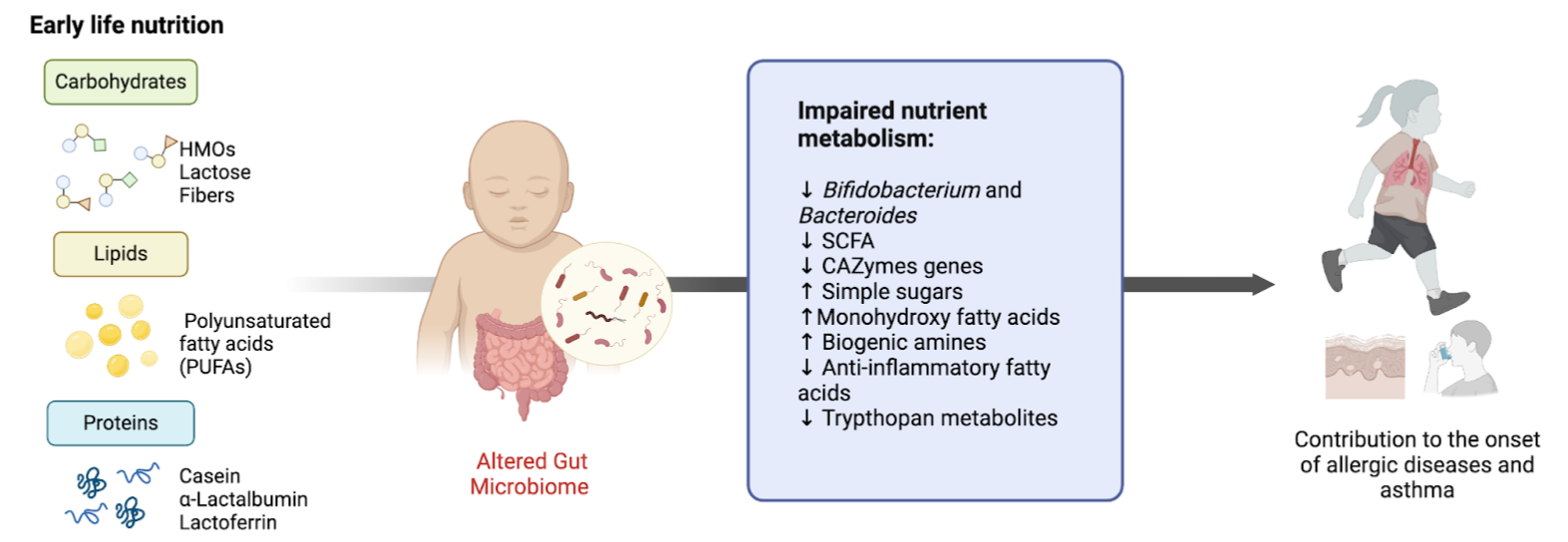
To investigate these dynamics, we cultivate in vitro microbial communities derived from infant fecal samples, representing both healthy infants and those who later develop asthma. These models enable the controlled study of how dietary components, reflective of early-life nutrition, influence microbial composition and their inflammatory potential. By employing diverse media formulations, we aim to uncover how specific nutrients modulate microbial structure and functionality, shedding light on the intricate interplay between diet, gut microbial communities, and immune responses during early life.
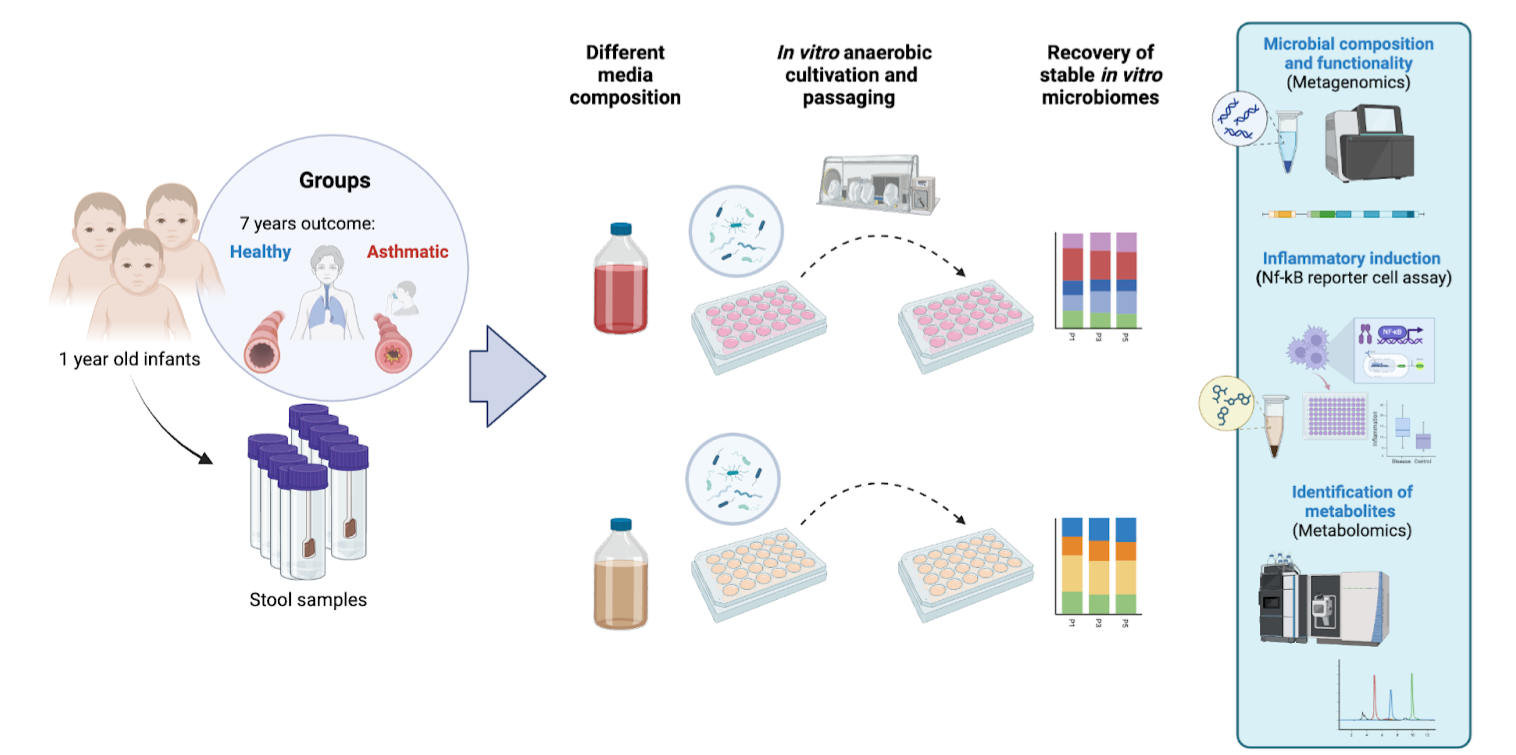
This research provides a platform to explore the mechanistic underpinnings of microbiome-driven immune responses and identify functional microbial targets for with therapeutical potential. By bridging experimental microbiology with systems biology, our work aspires to inform innovative strategies for personalized microbiome modulation in early childhood that influence the development of allergic diseases such as asthma.