Plasmidomics
Overview
In situ microbiome editing will rely on the delivery and maintenance of heterologous (i.e. introduced) DNA in target cells. Plasmids are self-replicating genetic elements that propagate between microbial hosts and are commonly used in biotechnology as genetic vectors. While extensive research has focused on plasmids of clinically relevant bacteria, plasmid diversity, replication machinery, and functions in natural environments are vastly underexplored. Furthermore, current plasmid-based genome editing toolsets are functional only in select model microorganisms, limiting the ability to transform the uncultivated majority of species. A comprehensive characterization of rumen plasmids and their host relationships, undertaken by our lab, will lay the groundwork to harness this naturally-occurring DNA delivery modality to engineer the rumen microbiome to disrupt methane production at the source.
Research Areas and Approaches
We are conducting a systematic investigation of plasmid diversity and microbial host associations in the cow rumen. While mining of public sequence data has substantially expanded known plasmid diversity across biomes, current models are trained on relatively limited plasmid diversity known mainly from microbial isolates. Our approach is to examine circular elements in cow rumen metagenomes, with the potential to discover novel plasmids of uncultivated species. Detecting plasmids of methanogenic archaea—the microbes directly responsible for enteric emissions—is a core focus as knowledge of plasmids in these lineages is severely limited.
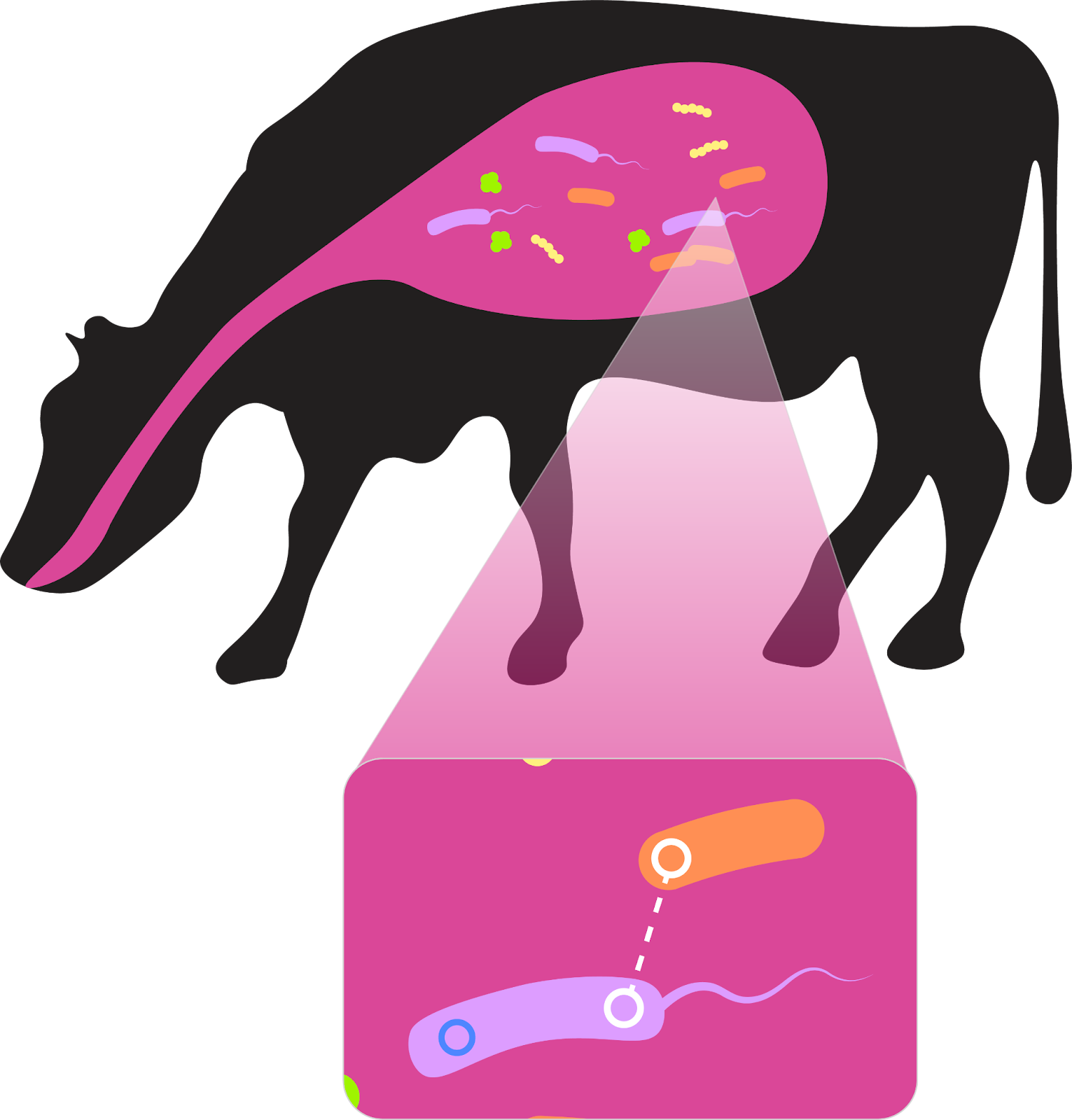
Associating plasmids and hosts using cultivation-independent approaches at community scale is highly challenging. We are associating plasmids and hosts using intracellular proximity ligation and high-throughput chromosome conformation capture (Hi-C; illustrated below). Statistical methods developed in the lab are critical for denoising the data. Informatic plasmid–host predictions will be validated in in vitro systems with the help of our IGI collaborators.
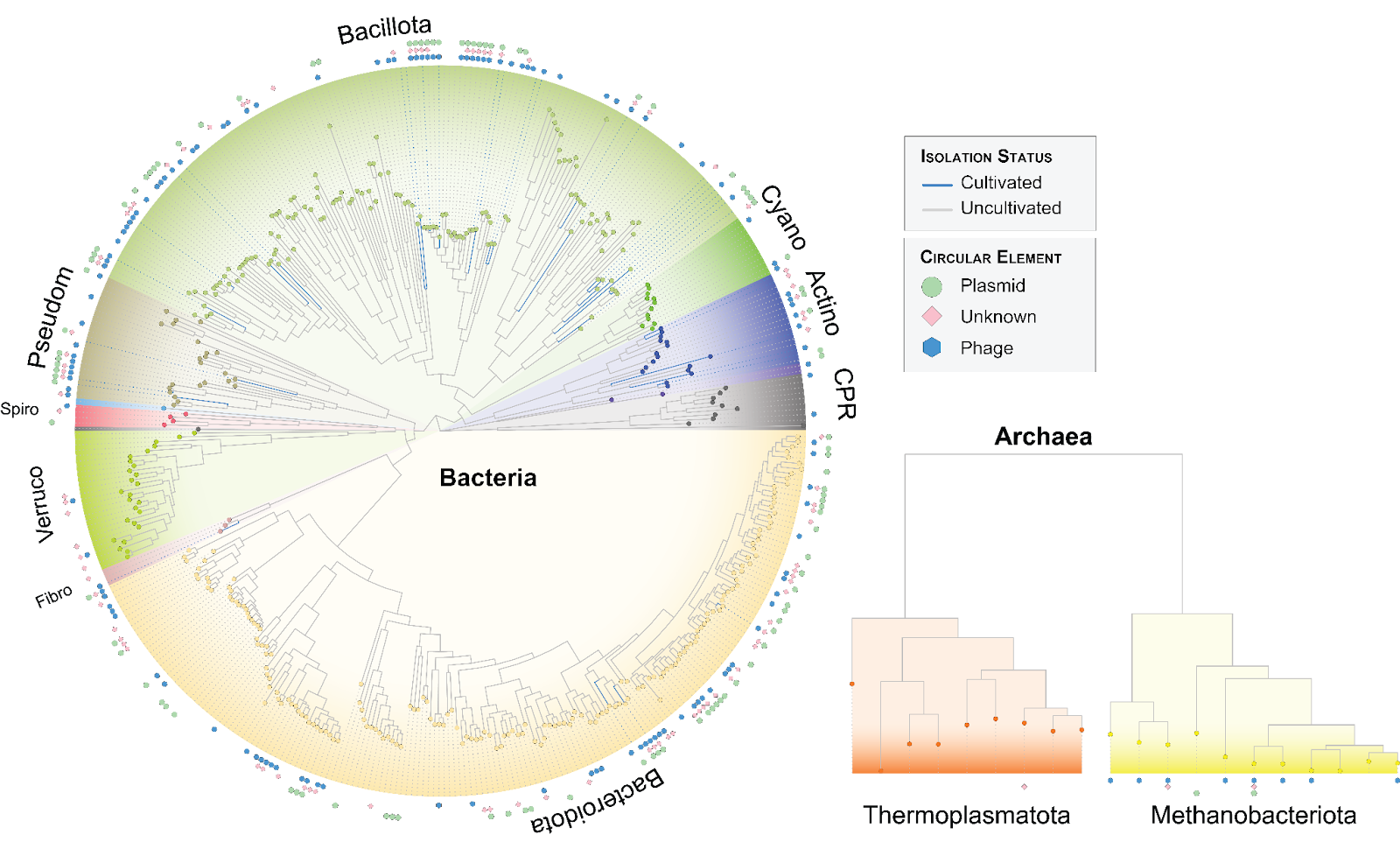
 Hi-C can recover plasmid-host associations by (1) chemically cross-linking the chromosome (orange) and plasmid (green) based on their physical proximity within a cell. (2) The DNA of each molecule is fragmented at restriction sites and the ends are filled and biotinylated. (3) Chromosome and plasmid fragments are ligated and (4) sheared to generate chimeric molecules, which are enriched by pulling down the biotin. (5) Paired-end sequencing is performed on the chimeric chromosome-plasmid fragments. Adapted from
Hi-C can recover plasmid-host associations by (1) chemically cross-linking the chromosome (orange) and plasmid (green) based on their physical proximity within a cell. (2) The DNA of each molecule is fragmented at restriction sites and the ends are filled and biotinylated. (3) Chromosome and plasmid fragments are ligated and (4) sheared to generate chimeric molecules, which are enriched by pulling down the biotin. (5) Paired-end sequencing is performed on the chimeric chromosome-plasmid fragments. Adapted from

We are also examining replication machinery allowing plasmids to replicate in a range of microbial hosts. So far, replication systems have not been broadly characterized. Elucidating replicon diversity including metabolic gene clusters will highlight replicative units to edit a broad diversity of rumen species. In collaboration with the Sachdeva Lab, we are examining gene clusters in replication and transfer origins (oriV and oriT) predicted by large language models with the capability to detect novel plasmid features. In vitro experimentation performed in collaboration with other IGI labs will test the functionality of informatically predicted replicons.